The incredible diversity of microbial life is not only driven by the different genes in their genomes,
but also by the chemical structure of the DNA which encodes those genes – their “epigenome.”
The Pan-Epigenome Browser makes it easy to compare the epigenetic modifications from a group of
organisms, identifying important patterns to learn something new about the microbial world.
Example Datasets
| Organism | Browser | Data Files | Num. Genomes | Num. Motifs |
|---|---|---|---|---|
| None | link | N/A | 0 | 0 |
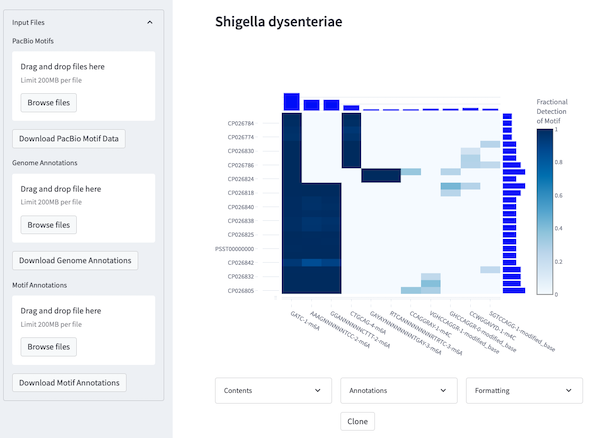
| Shigella dysenteriae | link | link | 26 | 44 |
| Escherichia coli | link | link | 103 | 389 |
| Clostridioides difficile | link | link | 40 | 34 |
| Streptococcus equi subsp. equi | link | link | 35 | 20 |
| Klebsiella pneumoniae | link | link | 31 | 95 |
Background
There are many different reasons why epigenetic modifications may be important for the biology
of a particular bacterial strain:
- Defense from invading genetic elements (e.g. plasmids, phage)
- Regulation of gene expression
- Phase variation
The large amount of variation in bacterial epigenetics has become much better appreciated with the advent of third-generation epigenome-aware platforms for genome sequencing. The primary platform which provides epigenetic information is PacBio SMRT Sequencing, while the Oxford Nanopore platform can also generate methylation information
Some key characteristics of bacterial epigenetic modification are:
- Chemical modifications are made to DNA in the context of short nucleotide motifs (e.g. GATC)
- Each type of modification may occur at a different molecular position within the nucleotide
- Different strains of the same species will often have a distinct epigenetic profile (collection of motifs)
Input Data
The Pan-Epigenome Browser reads epigenome profiles which are produced by the PacBio analysis platform.
Specifically, the motifs.csv file produced by the
MotifMaker
utility for each genome.
It is strongly recommended to name each of the motif CSV files according to their source to avoid
confusion (e.g. EcoliK12.motifs.csv).
Public Data
The public NCBI data repository
accepts PacBio base modification files
including the summary CSV format which is used by the Pan-Epigenome Browser.
A complete list of datasets which include these files can be found
here.
Within that table, any of the files with BaseModification-MotifsSummary listed
as their File Type should be compatible.
In addition, the BioProjects containing base modification data can be found using
the search term
“has basemodification”[Properties]
Background Reading
Wilbanks, E.G., Doré, H., Ashby, M.H. et al. Metagenomic methylation patterns resolve bacterial genomes of unusual size and structural complexity. ISME J 16, 1921–1931 (2022). https://doi.org/10.1038/s41396-022-01242-7

Fig. 1: Restriction-modification (RM) systems provide bacteria and archaea with a defense against foreign DNA by discriminating self from non-self DNA based on methylation patterns.